How to save (and load) datasets in R: An overview
What I will show you
In this post, I want to show you a few ways how you can save your datasets in R. Maybe, this seems like a dumb question to you. But after giving quite a few R courses mainly - but not only - for R beginners, I came to acknowledge that the answer to this question is not obvious and the different possibilites can be confusing. In this post, I want to give an overview over the different alternatives and also state my opinion which way is the best in which situation.
Why would you want to know that?
Well, there are quite a few tutorials out there on how to read data into R. RStudio even has a special button for this in the ‘Environment’ tab - it’s labelled ‘Import Dataset’. But there is no button and also fewer tutorials on saving data. That’s strange, isn’t it? If you import your data, you might do some (sometimes lengthy) manipulation, aggregation, selection and other stuff. If all that stuff takes several minutes (or even longer), you might not want to do it everytime you are working with the data. So, you might want to save your dataset at a stage that’s pre-analyses but post-processing (where ‘processing’ might include cleaning, manipulating, calculating new variables, merging, selecting, aggregating and lots of other stuff).
What are we going to do?
I will show you the following ways of saving or exporting your data from R:
- Saving it as an R object with the functions
save()andsaveRDS() - Saving it as a CSV file with
write.table()orfwrite() - Exporting it to an Excel file with
WriteXLS()
For me, these options cover at least 90% of the stuff I have to do at work. So I hope that it’ll work for you, too.
Preparation: Load some data
I will use some fairly (but not very) large dataset from the car package. The dataset is called MplsStops and holds information about stops made by the Minneapolis Police Department in 2017. Of course, you can access this dataset by installing and loading the car package and typing MplsStops. However, I want to simulate a more typical workflow here. Namely, loading a dataset from your disk (I will load it over the WWW). The dataset is also available from GitHub:
data <- read.table("https://vincentarelbundock.github.io/Rdatasets/csv/carData/MplsStops.csv",
sep = ",", header = T,
row.names = 1)
scroll_box(kable(head(data), row.names = F),
width = "100%", height = "300px")| idNum | date | problem | MDC | citationIssued | personSearch | vehicleSearch | preRace | race | gender | lat | long | policePrecinct | neighborhood |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 17-000003 | 2017-01-01 00:00:42 | suspicious | MDC | NA | NO | NO | Unknown | Unknown | Unknown | 44.96662 | -93.24646 | 1 | Cedar Riverside |
| 17-000007 | 2017-01-01 00:03:07 | suspicious | MDC | NA | NO | NO | Unknown | Unknown | Male | 44.98045 | -93.27134 | 1 | Downtown West |
| 17-000073 | 2017-01-01 00:23:15 | traffic | MDC | NA | NO | NO | Unknown | White | Female | 44.94835 | -93.27538 | 5 | Whittier |
| 17-000092 | 2017-01-01 00:33:48 | suspicious | MDC | NA | NO | NO | Unknown | East African | Male | 44.94836 | -93.28135 | 5 | Whittier |
| 17-000098 | 2017-01-01 00:37:58 | traffic | MDC | NA | NO | NO | Unknown | White | Female | 44.97908 | -93.26208 | 1 | Downtown West |
| 17-000111 | 2017-01-01 00:46:48 | traffic | MDC | NA | NO | NO | Unknown | East African | Male | 44.98054 | -93.26363 | 1 | Downtown West |
We now have a dataset with over 50,000 rows (you can scroll through the first 6 of them in the box above) and 14 variables in our global environment (the ‘workspace’). Just for the sake of simulating a real workflow, I will do some very light data manipulation. Here, I’m assigning a new column data$gender.not.known which is TRUE whenever data$gender is "Unknown" or NA.
data$gender.not.known <- is.na(data$gender) | data$gender == "Unknown"As I wrote above: Saving the current state of your dataset in R makes sense when all the preparations take a lot of time. If they don’t, you can just run your pre-processing code every time you are getting back to analyzing the dataset. In the scope of this post, let’s suppose that the calculation above took veeeery long and you absolutely don’t want to run it everytime.
Option 1: Save as an R object
Whenever I’m the only one working on a project or everybody else is also using R, I like to save my datasets as R objects. Basically, it’s just saving a variable/object (or several of them) in a file on your disk. There are two ways of doing this:
- Use the function
save()to create an.Rdatafile. In these files, you can store several variables. - Use the function
saveRDS()to create an.Rdsfile. You can only store one variable in it.
Option 1.1: save()
You can save your data simply by doing the following:
save(data, file = "data.Rdata")By default, the parameter compress of the save() function is turned on. That means that the resulting file will use less space on your disk. However, if it is a really huge dataset, it could take longer to load it later because R first has to extract the file again. So, if you want to save space, then leave it as it is. If you want to save time, add a parameter compress = F.
If you want to load such an .Rdata file into your environment, simply do
load(file = "data.Rdata")Then, the object is available in your workspace with its old name. Here, the new variable will also have the name data. With save() You can also save several objects in one file. Let’s duplicate data to simulate this.
data2 <- data
save(list = c("data", "data2"), file = "data.Rdata")Now, if you do load("data.Rdata"), you will have two more objects in your workspace, namely data and data2.
Option 1.2: saveRDS()
This is the second option of saving R objects. saveRDS() can only be used to save one object in one file. The “loading function” for saveRDS() is readRDS(). Let’s try it out.
saveRDS(data, file = "data.Rds")
data.copy <- readRDS(file = "data.Rds")Now, you have another R object in your workspace which is an exact copy of data. The compress parameter is also available for readRDS().
Note that you cannot “mix” the saving and loading functions: save() goes together with load(), saveRDS() goes together with readRDS().
The difference between save() and saveRDS()
So, you might ask “why should I use saveRDS() instead of save()”? Actually, I like saveRDS() better - for one specific reason that you might not have noticed in the calls above. When we use load(), we do not assign the result of the loading process to a variable because the original names of the objects are used. But this also means that you have to “remember” the names of the previously used objects when using load().
When we use readRDS(), we have to assign the result of the reading process to a variable. This might mean more typing but it also has the advantage that you can choose a new name for the variable to integrate it in into the rest of the new script more smoothly. Also, it is more similar to the behavior of all the other “reading functions” like read.table(): for these, you also have to assign the result to a variable. The only advantage of save() really is that you can save several objects into one file - but in the end it might be better to have one file for one object. This might be more clearly organized.
Option 2: Save as a CSV file
Whenever you are not so who will work with the data later on and whether these people are all using R, you might want to export your dataset as a CSV file. Also, it’s human readable. Also, if you provide a dataset on some website (e.g. in the Dataverse for other researchers, it is kind to provide a CSV file because everyone can open it with their preferred statistical software package.
Option 2.1: write.table()
You can think of write.table() as the “opposite” of read.table(). Even the parameters are quite similar. Let’s try it:
write.table(data, file = "data.csv",
sep = "\t", row.names = F)We just saved the data.frame stored in data as a CSV file with tabs as field separators. We also suppressed the rownames. I don’t know why, but by default, write.table() is storing the rownames in the file which I find a little strange.
Oh, and you can also use write.table() to append the contents of your data.frame at the end of the file: just set the parameter append to TRUE. This is great whenever you want to “fill” a file in multiple steps (e.g., in a for loop). Remember to suppress the column names if you’re appending content to files because you don’t want them to be repeated throughout the file - simply set col.names = F.
Option 2.2: fwrite()
Is your dataset really huge, like several gigabytes of data? Then try giving fwrite() from the data.table package a spin! It uses multiple CPU cores for writing data. Just like fread() from the same package, it is much much faster for larger files. Another advantage: the row.names parameter is FALSE by default. The most-widely used parameters have the same names as in write.table(). Neat!
library(data.table)
t0 <- Sys.time()
for (i in 1:10) {
write.table(data,
file = "writetable.data.csv",
sep = "\t", row.names = F) }
difftime(Sys.time(), t0)## Time difference of 2.853841 secst1 <- Sys.time()
for (i in 1:10) {
fwrite(data,
file = "fwrite-data.csv",
sep = "\t") }
difftime(Sys.time(), t1)## Time difference of 0.3355439 secsSee that? Even with only 10 replications of writing a rather small dataset to disk, fwrite() has a huge timing advantage (it’s more than 10 times faster!). For a very large dataset, this might come in really handy.
Option 3: Save as an Excel file
You might come into a situation where you want to export your dataset to an Excel file. Maybe some colleagues only work with Excel (because you still not managed to convince them switching to R) or you want to use Excel for annotating your dataset with a spreadsheet editor. In this case, you can use the Write.XLS() function from the Write.XLS package. I tried a few packages for writing Excel files and I find this one the most convenient to use.
Let’s try it.
library(WriteXLS)
WriteXLS(data, ExcelFileName = "data.xlsx",
SheetNames = "my data",
AdjWidth = T,
BoldHeaderRow = T)This is what the resulting Excel file looks like on my machine.
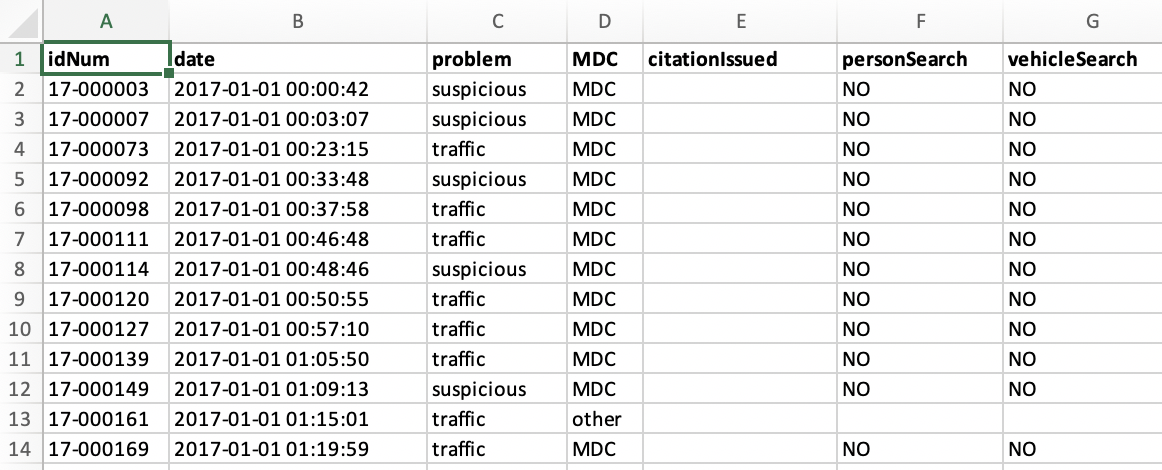
You can save several dataframes in one Excel file by including the names of the objects at the first position. Here, you could replace data with c("data", "data2"). With the parameter SheetNames you can set the names of the data sheets (visible at the bottom of Excel, not included in the screenshot). If you want to write several data.frames into several sheets of the Excel file, you can put several names in a vector here that have to correspond with the names of the objects at the first position.
Adj.Width is a nice parameter because it tries to adjust the width of the columns in Excel in a way that every entry fits in the cells. BoldHeaderRow is self-explanatory, I guess. You can see the effect in the screenshot. Oh, and by the way, you can set the entries for NA values with the na parameter. It’s "" by default.
Summary
- If you know that the dataset is going to be used in R and R only, use
saveRDS().save()is OK, too. But you cannot assign the result ofload()ing your data back into R to a variable name of your choice. You can save uncompressed files by settingcompress = F. Reading those files back in is much faster but they use more space on your disk. - If you want to distribute your dataset to a lot of people from whom you don’t know which statistical processing software package they use, you can save CSV files. I recommend using
fwrite()from thedata.tablepackage because it is much faster thanwrite.table(). - If you really really want (or need) an Excel file, I recommend using
WriteXLS()from theWriteXLSpackage.
If you think that I should also cover other formats of saving a dataset on the disk, please let me know in the comments and I will try to cover them as well.

